10 rows BEFORE YOU START. Extending the phyloseq class.
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
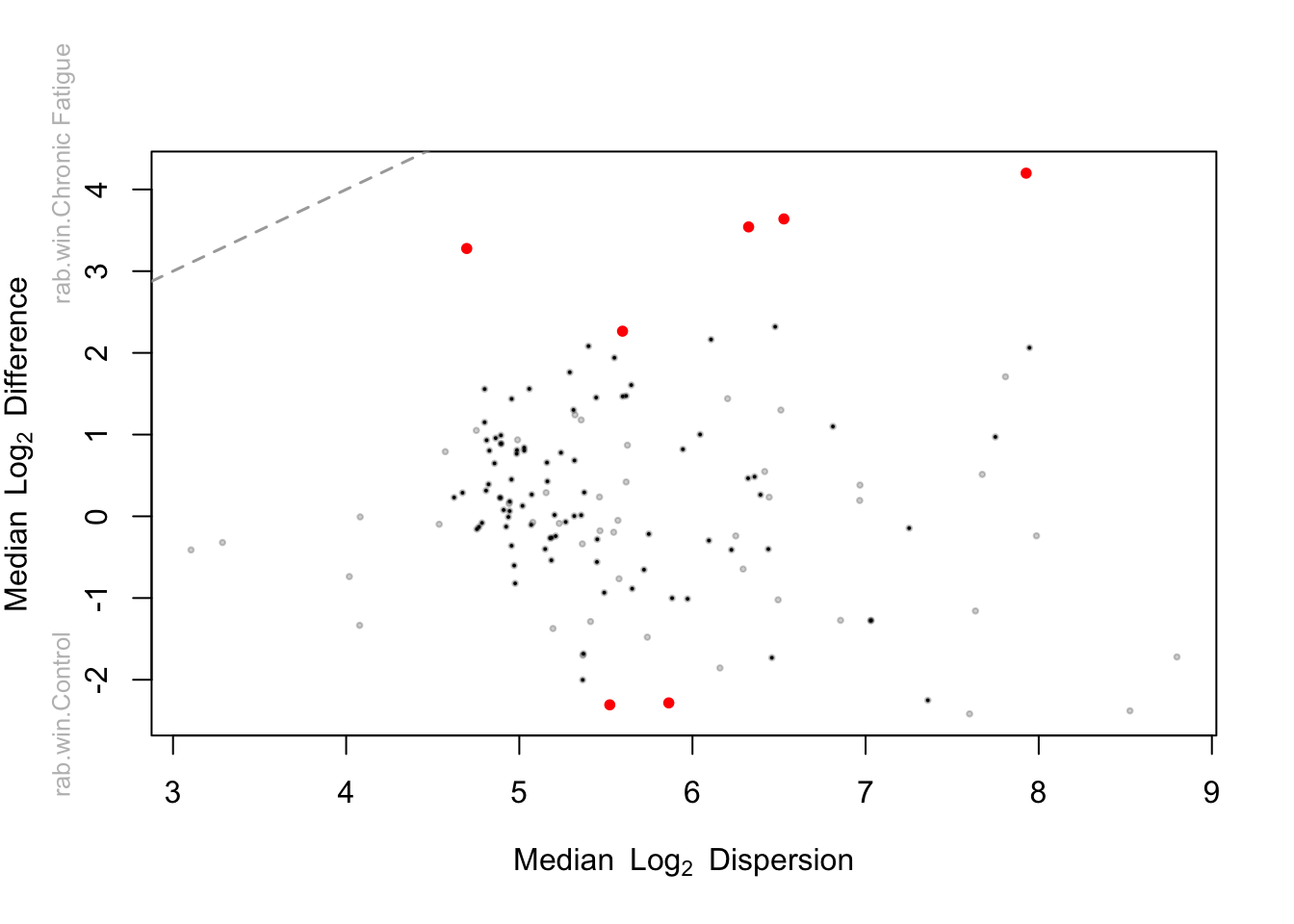
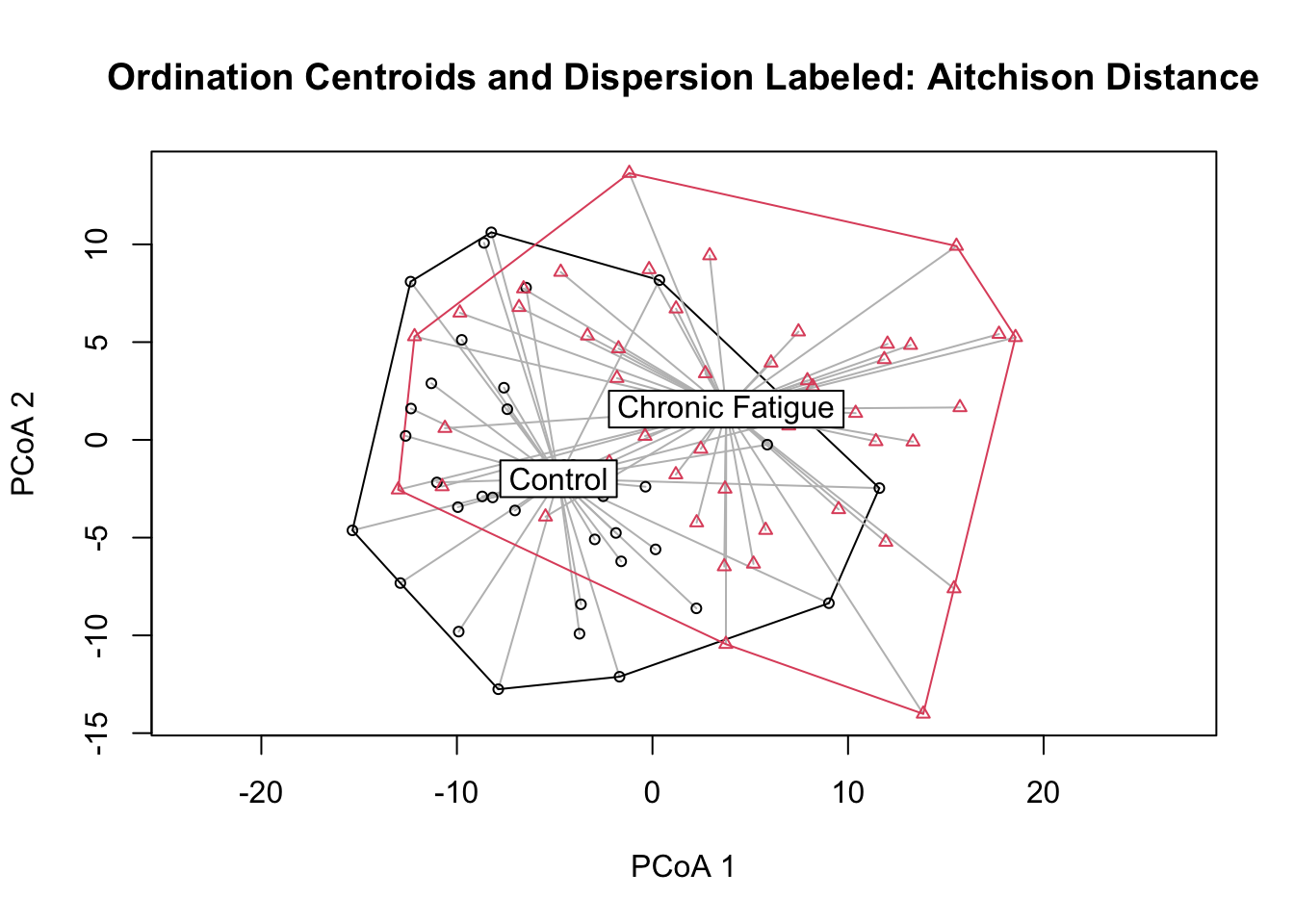
Beta diversity measures changes in species diversity.
. The package is in Bioconductor and aims to provide a comprehensive collection of tools and tutorials with a particular focus on amplicon sequencing data. Up to 10 cash back All the packages discussed in this method come with manual or tutorials which we encourage the users to read carefully for more information about the available methods their applications and detailed real-life or toy examples. Last updated almost 2 years ago.
We provide standard tools as well as novel extensions on standard analyses to improve interpretability and the analysts ability to communicate results all while maintaining object malleability to encourage open source collaboration. Using network theory one can model and analyze a microbiome and all its complex interactions in a single network. By Nabiilah Ardini Fauziyyah.
Metacoder has functions for parsing specific file formats used in metagenomics research. A reliable alternative to popular microbiome analysis R packages. This primer provides a concise introduction to conducting applied analyses of microbiome data in R.
The analysis of microbial communities brings many challenges. Tools for microbiome analysis. Helvetica-infected and pathogen-free DNA samples were used for 16S rRNA amplicon sequencing and microbiome analysis.
A tutorial on how to use Plotlys R graphing library for microbiome data analysis and visualization. Moving microbes from the 50 percent human artscience project. The microbiome R package facilitates phyloseq-based exploration and analysis of taxonomic profiling data.
If you are a moderator please see our troubleshooting guide. Explore microbiome profiles using R. In this all day workshop with lecture and hands on exercises.
Microbiome Analysis in R. 2013 OKeefe et al. Universidad de los Andes Bogota Colombia 3-7 December 2018.
The purpose of this tutorial is to describe the steps required to perform. This material has sense been taught at. Hide Comments Share Hide Toolbars.
2014 Lahti et al. A more comprehensive tutorial is available on-line. I see that the ordination plot of Unweighted unifrac suggests not much difference in microbiome in terms of composition.
The workshop is Part II of the microbiota analysis. In Part I we processed 16S-derived data and classified reads in operational taxonomic units OTUs with the software mothur. Lets continue to the next step of a typical microbiome analysis by looking at the beta diversity.
And Analyzing Microbiome Networks in R. This can be difficult for taxonomic data since it has a hierarchical component ie the taxonomic tree. The tutorial starts.
The sequencing reads have to be denoised and. Eds Microbiome Analysis. Many tools exist to quantify and compare abundance levels or OTU composition of communities in different conditions.
We will now take the results and go over the statistical analysis with the software R on a Linux server platform. High-throughput sequencing of PCR-amplified taxonomic markers like the 16S rRNA gene has enabled a new level of analysis of complex bacterial communities known as microbiomes. 3 years ago.
2015A more comprehensive tutorial is available on-line. The Rmarkdown source code html for all tutorials is available in the Github indexpage. This vignette provides a brief overview with example data sets from published microbiome profiling studies Lahti et al.
Tutorials for microbiome analysis - GitHub Pages. Thanks to Joey McMurdie joey711 Ben Callahan benjjneb and Mike Mclaren mmclaren42 for assisting with the original preperation of materials. But a PERMANOVA of unweighted unifrac says that samples are significantly different.
Beiko R Hsiao W Parkinson J. The data itself may originate from widely different sources such as the microbiomes of. With multiple example data sets from published studies.
While this primer does not require extensive knowledge of programming in R the user is expected to install R and all packages required for this primer. The first step in any analysis is getting your data into R. An R package for microbial community analysis in an environmental context.
However for this demonstration we will be using a more all-purpose parser from the taxa. Here we describe in detail and step by step the process of building analyzing and visualizing microbiome networks from operational taxonomic unit OTU tables in R and RStudio using several different approaches and extensively. Microbiome Analysis Using R.
Please install the required software and download the example data before coming to the workshop. Recently developed culture-independent methods based on high-throughput sequencing of 16S18S ribosomal RNA gene variable regions and internal transcribed spacers ITS enable researchers to identify all the microbes in their complex habitats or in other words to analyse a microbiome. Downstream plotting and analysis of 16s microbiome data in R using phyloseq and ggplot This beginner-friendly tutorial will allow you to create publication-level graphs and convert phyloseq objects into dataframes for easier manipulation and analysis.
The integration of many different types of data with methods from ecology genetics phylogenetics network analysis visualization and testing. Tutorial For Microbiome Analysis In R Yan Hui Below you will find R code for extracting alpha diversity beta diversity and taxa abundance. Below you will find R code for extracting alpha diversity beta diversity and taxa abundance.
Introduction to the microbiome R package. This is a tutorial to analyze microbiome data with R. While we continue to maintain this R package the development has been discontinued as we have.
This vignette provides a brief overview with example data sets from published microbiome profiling studies. The microbiome R package facilitates exploration and analysis of microbiome profiling data in particular 16S taxonomic profiling.

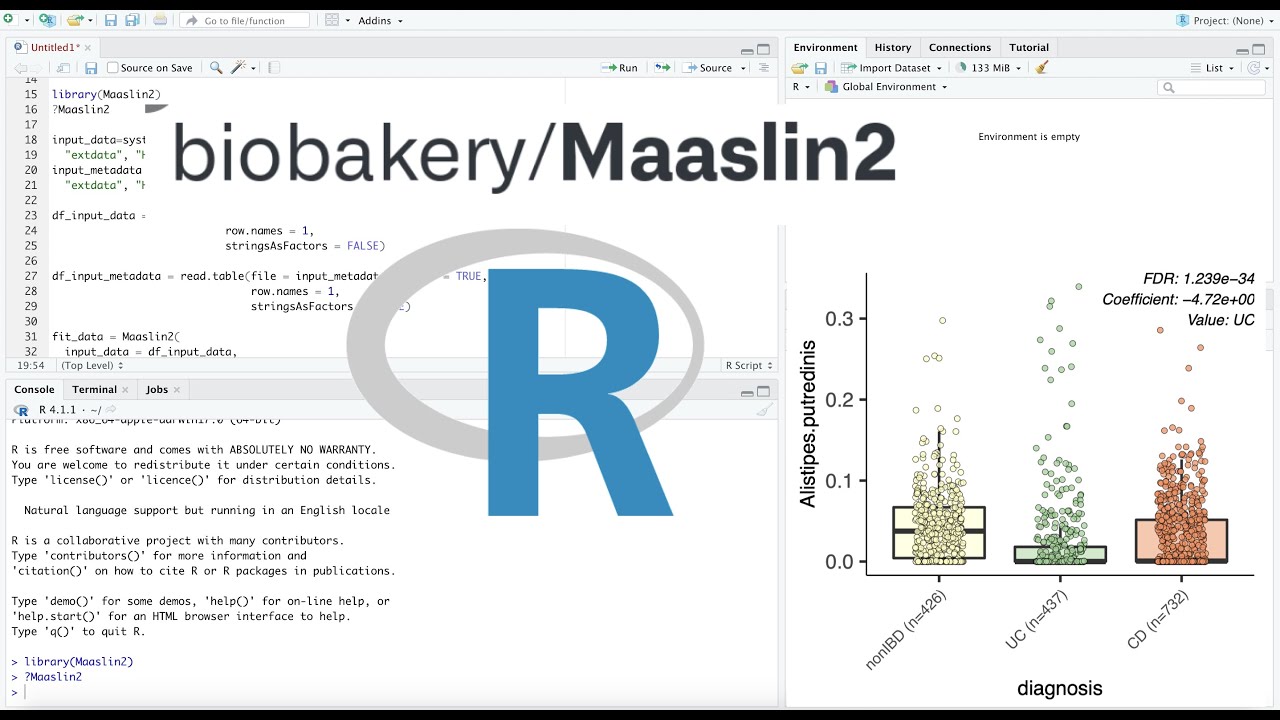
Maaslin2 Tutorial In R Studio Microbiome Multivariable Analysis Nutribiomes Youtube
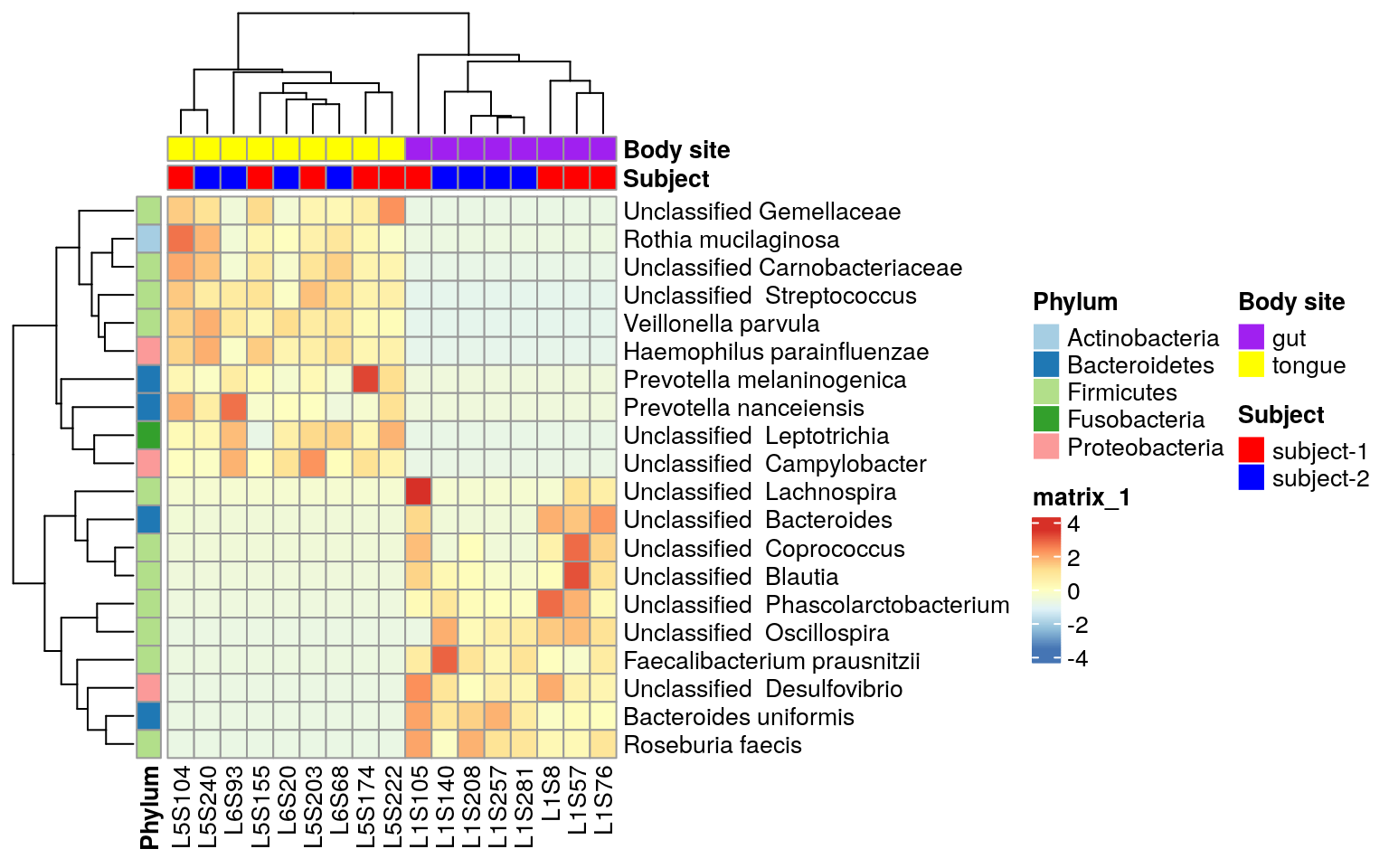
Tutorial For Microbiome Analysis In R Yan Hui

Making Heatmaps With R For Microbiome Analysis The Molecular Ecologist
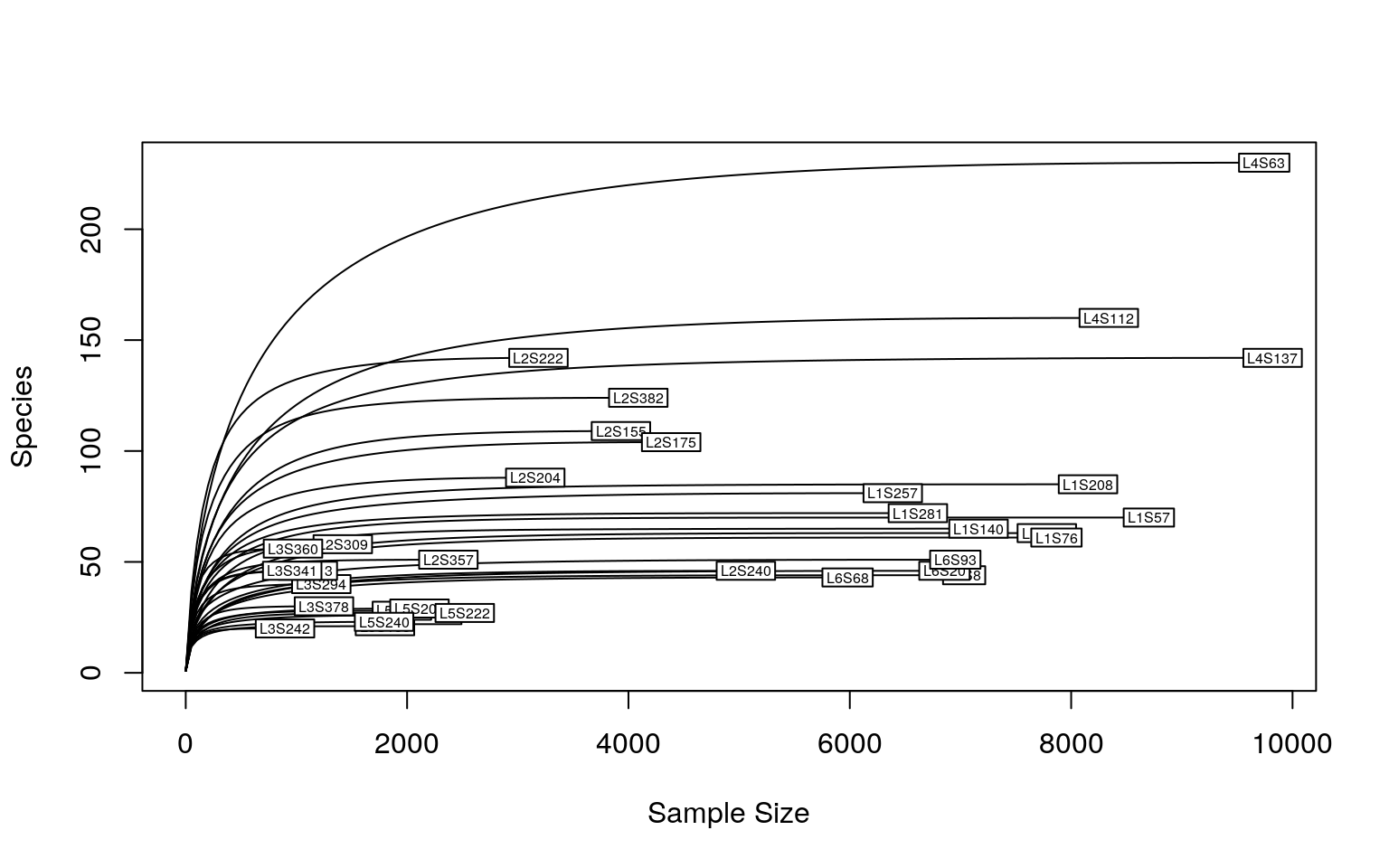
Tutorial For Microbiome Analysis In R Yan Hui
Introduction To The Statistical Analysis Of Microbiome Data In R Academic

0 comments
Post a Comment